Walkthrough on command line based tilt series alignment
This is the command line based version of the GUI based alignment walkthrough
Contents
Create the workflow
name = 'hivCommandLine'; folder = 'workflows'; u = dtsa(name,'--nogui','-path',folder,'fp',1);
Here, we instruct Dynamo to skip opening the GUI. We also create the workflow in a different folder.
Entering the data
The u object contains several areas to interact with the workflow. They can be found by autocompletion using the tab key. Here, we will proceed step by step; remember that you can write all the command lines in a single .m script.
Basic data
u.enter.tiltSeries(file);
u.enter.tiltAngles(-57:3:60);
If you want to reject some of the hight tilts (or any other view that appears to have been damages,)
u.enter.discardedTiltIndices([1,40]);
Acquisition settings
u.enter.settingAcquisition.apix(2.7);
Computation settings
u.enter.settingComputing.parallelCPUUse(1);
Detection settings
These here are the actual design decisions when running an alignment workflow:
u.enter.settingDetection.beadRadius(16);
u.enter.settingDetection.maskRadius(28);
u.enter.templateSidelength(64);
The bin level is mainly used to accelerate the detection procedure. Needs to be chosen in a way that a binned gold bead still can be recognisable as such, with a radius of at least 4 pixels.
u.enter.settingDetection.detectionBinningFactor(1);
Changing generic parameters
Yo can find handles to the parameters of the individuals steps through auto ompletion on the area, then step items.
u.area.indexing.step.tiltGapFiller.parameterSet.residualsThreshold(8);
Running the workflow
u.run.all('noctf',1);
On the fly reconstruction of particles
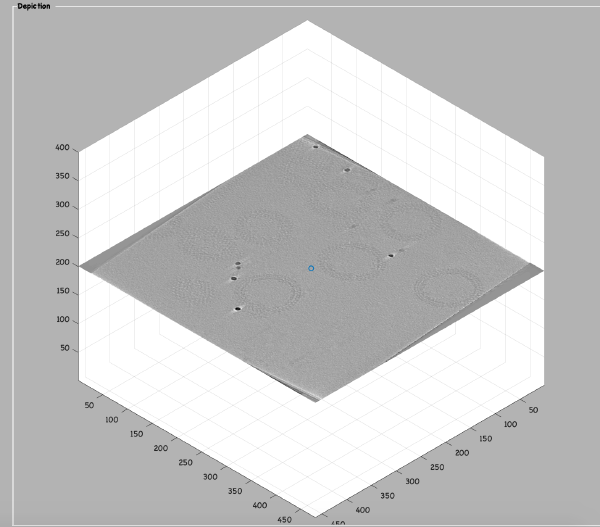
Grep coordinates in the binned tomogram
Open the binned reconstruction with dtmslice dtmslice 'workflows/hivCommandLine.AWF/reconstruction/binnedReconstructionWBP.mrc';
Now we can save the table as a file: write(temp_table,'binnedGoldbeads.tbl');
Cropping the gold bead particles
tomo = 'workflows/workshop.AWF/reconstruction/binnedReconstructionWBP.mrc';
tbl = 'binnedGoldbeads.tbl';
targetBinnedParticles = 'binnedGoldBeads.Data';
sidelength = 32;
o = dtcrop(tomo,tbl,target,sidelength);
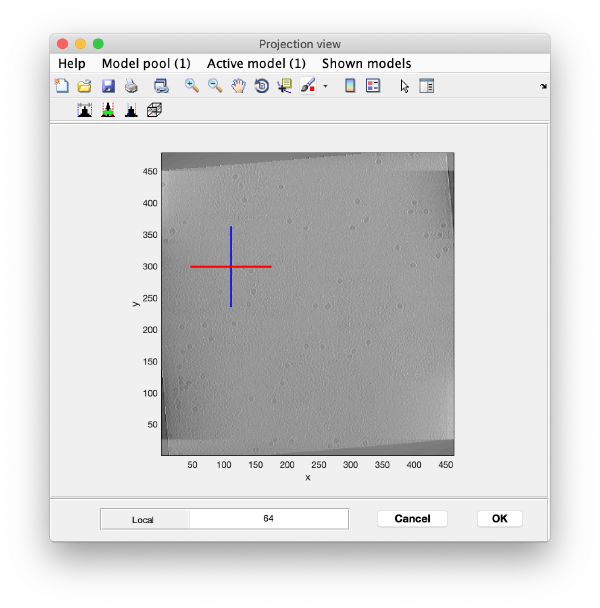
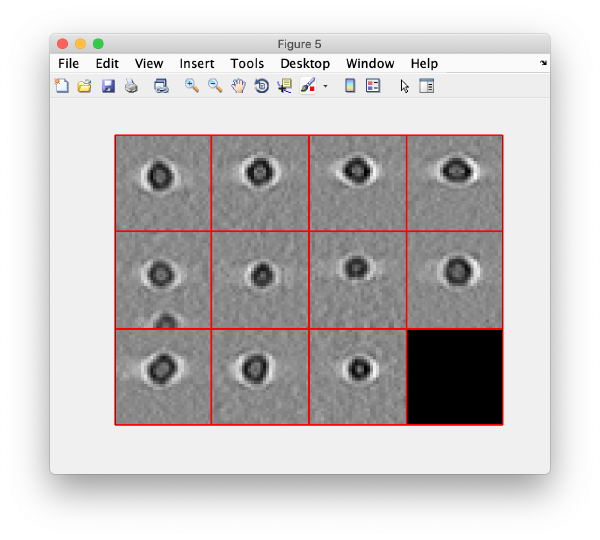
And then we visualise a projection along z of each cropped particle
dslices(targetBinnedParticles,'proj','c');shg;
Reconstruct particles on the fly
The command that reconstructs full sized particles from an aligned stack is dynama_table_trec. This is an independent function, not part of the workflow, implying that we will need to perform some format conversion.
tbl = dread('binnedGoldbeads.tbl');
We scale the table
alignmentBinLevel = 2;
tblFull = dynamo_table_rescale(tbl,'factor',2^alignmentBinLevel);
source = 'workflows/hivCommandLine AWF/align/alignedFullStack.mrc';
We create a data folder to contain the particles reconstructed on the fly.
targetDirectory = 'tempCrop.Data';
Angles actually used during the reconstruction are contained in the reconstructionTiltAngles' item of the output folder. Note that they might differ from the nominal angles, as some views might have been eliminated, both explicitely by the user, of by the program not being able to align all tilts.
angles = 'workflows/hivCommandLine.AWF/align/reconstructionTiltAngles.tlt';
The (binned) coordinates were measured in a binned tomogram, whose dimensionality is requested by dynamo_table_rec. We need to provide dynamo_table_rec with visualizationTomogram = 'workflows/ hivCommandLine.AWF/reconstruction/binnedReconstructionWBP.mrc'; sizeTomogram = dynamo_read_size(visualizationTomogram); sizeTomogramFull = sizeTomogram*2^alignmentBinLevel;
The tomogram center can be shifted in order to account for a possible shift in the centre in the binned reconstruction that was used to crop the particles.
shiftTomogramCenter = [0,0,0];
We proceed now to the reconstruction.
sidelengthFullSize = 128;
dynamo_table_rec(tblFull,source,angles,targetDirectory,sidelength,....
'applyRampFilter',1,....
'sizeTomogram',sizeTomogramFull,....
'shiftTomogramCenter',shiftTomogramCenter);
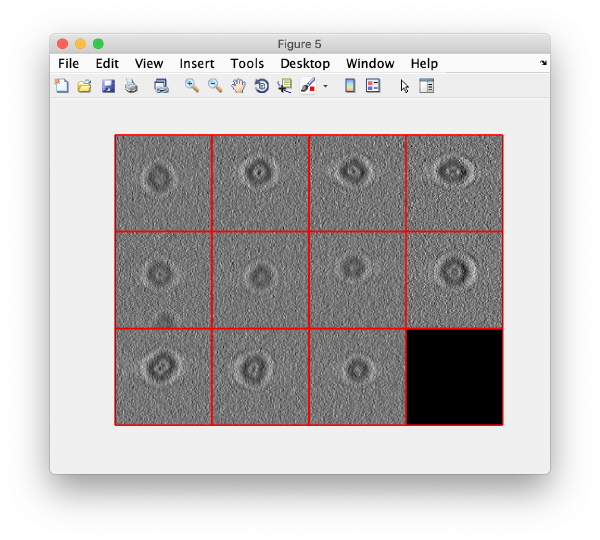
We can now check that the particles have been reconstructed in full size.
figure;dslices(targetDirectory,'proj','c');shg;